Predict#
Similar to training, prediction can be done via three interfaces:
via python,
das.predict.predictvia the command line,
das predict, with audio data from a wav file.the GUI - see the GUI tutorial
Prediction will:
load the audio data and the network
run inference to produce confidence scores (
class_probabilties)post-process the confidence score to extract the times of events and label segments.
Prediction using python#
import numpy as np
from pprint import pprint
import scipy.io.wavfile
import das.predict
help(das.predict.predict)
Help on function predict in module das.predict:
predict(x: numpy.ndarray, model_save_name: str = None, verbose: int = 1, batch_size: int = None, model: keras.engine.training.Model = None, params: dict = None, event_thres: float = 0.5, event_dist: float = 0.01, event_dist_min: float = 0, event_dist_max: float = None, segment_thres: float = 0.5, segment_use_optimized: bool = True, segment_minlen: float = None, segment_fillgap: float = None, pad: bool = True, prepend_data_padding: bool = True)
[summary]
Usage:
Calling predict with the path to the model will load the model and the
associated params and run inference:
`das.predict.predict(x=data, model_save_name='tata')`
To re-use the same model with multiple recordings, load the modal and params
once and pass them to `predict`
```my_model, my_params = das.utils.load_model_and_params(model_save_name)
for data in data_list:
das.predict.predict(x=data, model=my_model, params=my_params)
```
Args:
x (np.array): Audio data [samples, channels]
model_save_name (str): path with the trunk name of the model. Defaults to None.
model (keras.model.Models): Defaults to None.
params (dict): Defaults to None.
verbose (int): display progress bar during prediction. Defaults to 1.
batch_size (int): number of chunks processed at once . Defaults to None (the default used during training).
Larger batches lead to faster inference. Limited by memory size, in particular for GPUs which typically have 8GB.
Large batch sizes lead to loss of samples since only complete batches are used.
pad (bool): Append zeros to fill up batch. Otherwise the end can be cut.
Defaults to False
event_thres (float): Confidence threshold for detecting peaks. Range 0..1. Defaults to 0.5.
event_dist (float): Minimal distance between adjacent events during thresholding.
Prevents detecting duplicate events when the confidence trace is a little noisy.
Defaults to 0.01.
event_dist_min (float): MINimal inter-event interval for the event filter run during post processing.
Defaults to 0.
event_dist_max (float): MAXimal inter-event interval for the event filter run during post processing.
Defaults to None (no upper limit).
segment_thres (float): Confidence threshold for detecting segments. Range 0..1. Defaults to 0.5.
segment_use_optimized (bool): Use minlen and fillgap values from param file if they exist.
If segment_minlen and segment_fillgap are provided,
then they will override the values from the param file.
Defaults to True.
segment_minlen (float): Minimal duration in seconds of a segment used for filtering out spurious detections. Defaults to None.
segment_fillgap (float): Gap in seconds between adjacent segments to be filled. Useful for correcting brief lapses. Defaults to None.
pad (bool): prepend values (repeat last sample value) to fill the last batch. Otherwise, the end of the data will not be annotated because
the last, non-full batch will be skipped.
prepend_data_padding (bool, optional): Restores samples that are ignored
in the beginning of the first and the end of the last chunk
because of "ignore_boundaries". Defaults to True.
Raises:
ValueError: [description]
Returns:
events: [description]
segments: [description]
class_probabilities (np.array): [T, nb_classes]
class_names (List[str]): [nb_classes]
%%time
samplerate, x = scipy.io.wavfile.read('dat/dmel_song_rt.wav')
print(f"DAS requires [T, channels], but single-channel wave files are loaded with shape [T,] (data shape is {x.shape}).")
x = np.atleast_2d(x).T
events, segments, class_probabilities, class_names = das.predict.predict(x,
model_save_name='models/dmel_single_rt/20200430_201821',
verbose=2,
segment_minlen=0.02,
segment_fillgap=0.02)
DAS requires [T, channels], but single-channel wave files are loaded with shape [T,] (data shape is (35000,)).
/Users/clemens10/miniconda3/lib/python3.9/site-packages/keras/layers/core.py:1043: UserWarning: dss.tcn.tcn is not loaded, but a Lambda layer uses it. It may cause errors.
warnings.warn('{} is not loaded, but a Lambda layer uses it. '
2022-07-25 13:02:25.335763: I tensorflow/core/platform/cpu_feature_guard.cc:142] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: SSE4.1 SSE4.2 AVX AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-07-25 13:02:25.918851: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)
20/20 - 2s
CPU times: user 5.92 s, sys: 895 ms, total: 6.82 s
Wall time: 2.62 s
Outputs of predict#
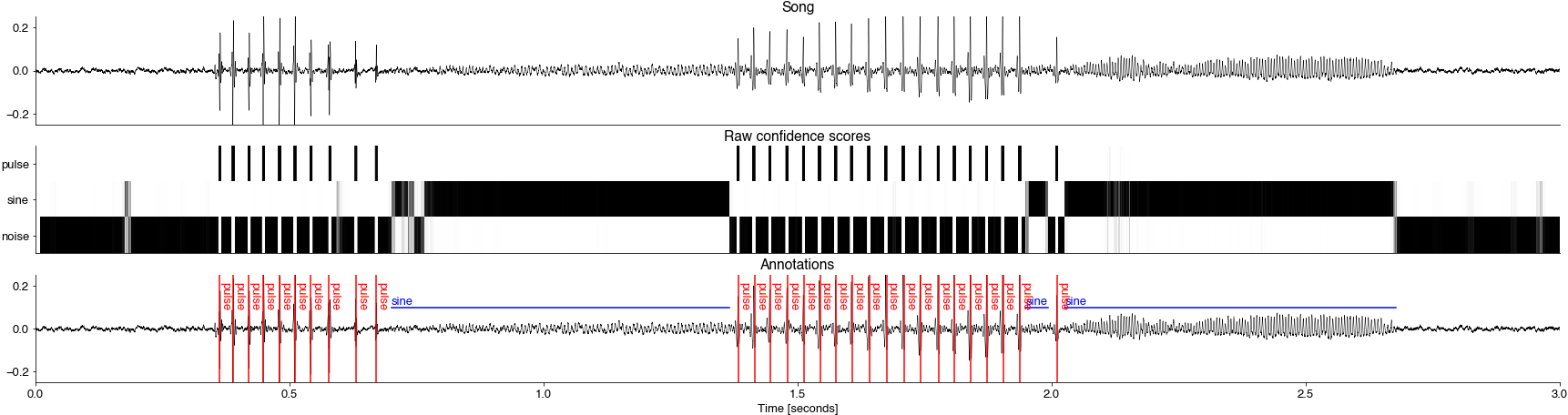
class_probabilties:[T, nb_classes]including noise.segments: Labelled segmentssamplerate_Hz:names: names of all segment typesindex: indices of all segments types into class_probabiltiiesprobabilities = class_probabilites[:, index]sequence: sequence of segment names (one entry per detected segment). Excludes noisesamples: labelled sample trace (label of the sequence occupying each sample)onsets_seconds,offsets_seconds,durations_seconds: Onsets, offsets, and duration of individual segmeents
events: Detected eventssamplerate_Hz:index: indices of all events types into class_probabiltiiesnames: names of all event typesprobabilities: probabilities (confidence scores) for detected events. Value ofclass_probabilitiesfor the detected event index at each event time.seconds: times (seconds) of detected eventssequence: sequence of event names (one per detected event).
import matplotlib.pyplot as plt
plt.style.use('ncb.mplstyle')
t0 = 0
t1 = 30_000
fs =segments['samplerate_Hz']
time = np.arange(t0, t1) / fs
nb_classes = class_probabilities.shape[1]
plt.figure(figsize=(30, 10))
plt.subplot(411)
plt.plot(time, x[t0:t1], 'k', linewidth=0.5)
plt.title('Song')
plt.xticks([])
plt.ylim(-0.25, 0.25)
plt.subplot(412)
plt.imshow(class_probabilities[t0:t1].T, cmap='Greys')
plt.yticks(np.arange(nb_classes), labels=class_names)
plt.title('Raw confidence scores')
plt.xticks([])
ax = plt.subplot(413)
plt.plot(time, x[t0:t1],'k', linewidth=0.5)
plt.ylim(-0.25, 0.25)
plt.title('Annotations')
plt.xlabel('Time [seconds]')
for onset, offset, segment_name in zip(segments['onsets_seconds'], segments['offsets_seconds'], segments['sequence']):
if onset >= t0 /fs and offset <= t1 / fs:
plt.plot([onset, offset], [0.1, 0.1], c='b')
ax.annotate(segment_name, xy=(onset, 0.11), c='b')
for pulse_time, pulse_name in zip(events['seconds'], events['sequence']):
if pulse_time >= t0 /fs and pulse_time <= t1 / fs:
plt.axvline(pulse_time, c='r')
ax.annotate(pulse_name, xy=(pulse_time, 0.1), c='r', rotation=-90)
Prediction using command-line scripts#
Will save the output of das.predict.predict to a h5 file ending in _das.h5 or specified via the --save-filename argument.
See cli for a full list of arguments.
!das predict dat/dmel_song_rt.wav models/dmel_single_rt/20200430_201821
INFO:root:Loading model from models/dmel_single_rt/20200430_201821.
2022-07-25 13:02:37.243085: I tensorflow/core/platform/cpu_feature_guard.cc:142] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: SSE4.1 SSE4.2 AVX AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
INFO:root: Loading data from dat/dmel_song_rt.wav.
INFO:root: Annotating using model at models/dmel_single_rt/20200430_201821.
2022-07-25 13:02:37.763758: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)
20/20 [==============================] - 2s 68ms/step
INFO:root: found 29 instances of events '['pulse']'.
INFO:root: found 15 instances of segments '['sine']'.
INFO:root: Saving results to dat/dmel_song_rt_annotations.csv.
INFO:root:Done.
import h5py
with h5py.File('dat/dmel_song_rt_das.h5', mode='r') as f:
print(list(f.keys()))
['class_names', 'class_probabilities', 'events', 'segments']